
How to access the MOSAIC dataset
Option 1 - Access via the EGA
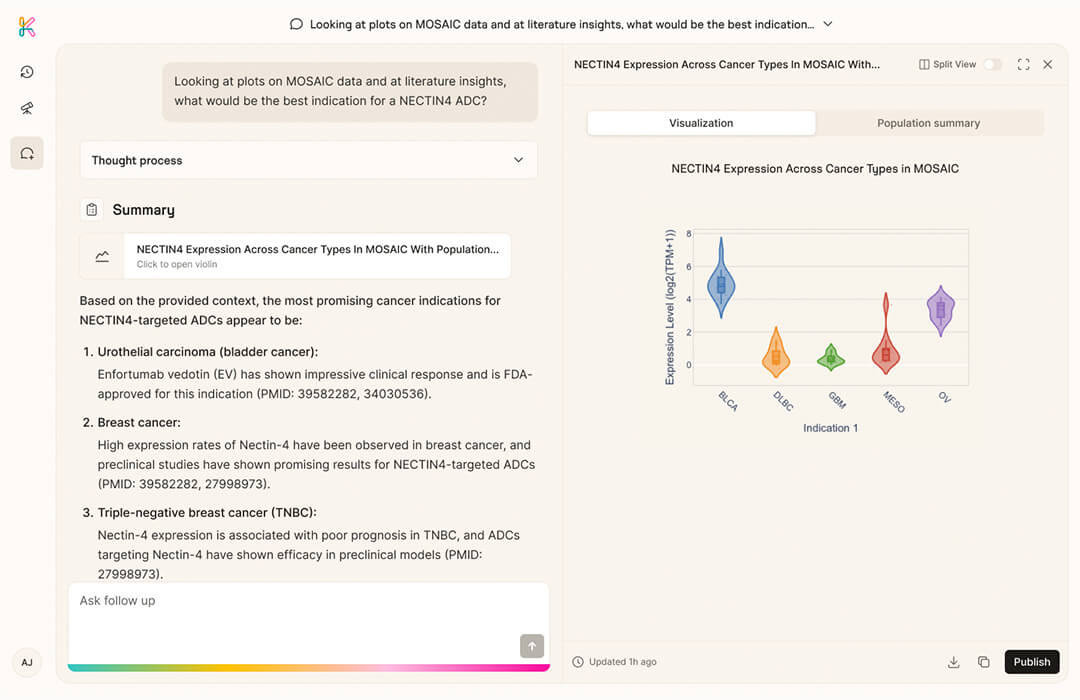
Option 2 - Explore via K Pro

The full MOSAIC dataset is available as an add on to the premium tier of K Pro. Talk to our team or book a demo to learn more about this option.

Large-scale spatial omics data is essential to fuel research in cancer
The convergence of large-scale spatial omics data and AI will power the next revolution of oncology research. There is an urgent need for the whole cancer community to have access to these data to reveal groups of patients with distinct tumour/immune biology.
The MOSAIC initiative will overcome this limitation by creating a large scale dataset of tumor spatial transcriptomes, allowing scientists to conduct research on data from cohorts larger than what is currently possible.
The MOSAIC dataset today:

Data modalities
MOSAIC covers six data modalities to capture the complexity of disease biology at multiple scales.
Here is a breakdown of current number of patient samples per modality in the dataset.
Cancer therapy areas
MOSAIC focuses on selected cohorts that include ten cancer indications.
Here is a breakdown of the number of patients per therapy area currently in the dataset

Why spatial biology?
Spatial biology brings a new dimension to cancer research because it maps biological molecules to their location within a tissue, capturing the spatial context.
When combined with data from technologies like imaging and genomics, spatial omics contributes to a rich, multiscale understanding of disease mechanisms.

How will MOSAIC help us fight cancer?
Through spatial omics MOSAIC will paint a picture of the dynamics between cells, highlighting key biological networks to enable precision medicine via the discovery of:
New disease biology
New patient subtypes
New biomarkers
New drug targets
Matching the right patient to the right treatment
Partners







